22 Kasım 2015 Pazar
Can you run Proteome Discoverer on Windows 10?
I've been asked this question a couple of times. And maybe now I know the answer.
At first run, it certainly looks like PD 2.1 installs just fine with nothing special whatsoever on Windows 10.
Now...that being said, this isn't officially supported by the vendor. And just because the couple HeLa runs I just did seem to go just fine doesn't mean that every feature will be ready...but, again, I haven't had a problem yet!
19 Kasım 2015 Perşembe
Nice and short review on proteogenomics in Springer
Like an awful lot of people right now, I'm kind of obsessed with the magical thing called "proteogenomics". Which...honestly...seems a little bit like magic. Getting good quality transcriptional data and filtering it so that you can see new mutations....that you can trust...AND THEN using this information to find new matches to your MS/MS data....that you can trust....
A few people have totally pulled it off....and I have the papers stacked up in front of my PC all marked up and highlighted and...well...maybe I'm dumb...but I still don't know how they did it.
For a good starting point, check out this nice review from Sam Faulkner et al.,. While I'm on the topic of magic, you might be surprised to see this is an article from Springer that isn't behind a paywall!
Oh yeah! And here is the link!
18 Kasım 2015 Çarşamba
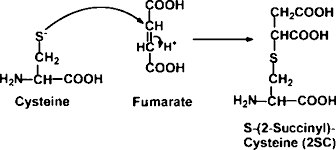
2SC --- another important PTM to look for?
Great...another protein post-translational modification...cause my search space isn't nutso enough already....
Oh! Hi, there! Ever looked at this one? Its called 2SC...and it turns out that it has a role in human diseases. Its even detectable by other molecular techniques and has been implicated in diabetes. A brand new paper from Gianluca Miglia et al., takes a computational approach and analyzes a ton of proteins. Turns out that it mostly sticks to Cysteines. Which makes me wonder if our typical techniques of cysteine reduction/alkylation/assumption of 100% cysteine alkylation means that we probably don't see it in our RAW data and can't see it in our processed data if it was there.
The abstract suggests that this modification is prevalent enough that they have seen multiple 2SC modifications per individual protein. I can't get much further because I'm stuck behind a paywall at this Panera this morning. An Open Access paper on this modification in people is available here, though its worth noting that they appear to have done all their work with Western blots. As a side note, it appears there are a lot of journals out there, and it appears that people are running out of names for them.
Ever wanted a great label free proteomics dataset?
"Hey guys, here's this awesome new algorithm for label free analysis!!!!"
Want a GREAT dataset to test it out on?
Check out this cool new paper from Claire Ramus et al.,! In this, they developed a great standard that all of us can download and use for free. They also use it to test a bunch of the current algorithms people are using.
The sample is the Sigma UPS1 48 protein mix (all equimolar proteins) spiked into a Yeast digest background at different concentrations. The spike-ins range from 50amol to 12,500 amol (yeah...there is a more efficient way of writing this, but I'm not great at metrics...thanks American public schools! I love sounding like an idiot to everyone else in the world...)
You can download the dataset...wow...my internet is slow right now...well, I guess you can download the dataset...from PRIDE. It will be PXD0001819 after it is published. The authors were kind enough to provide a way to download the dataset prior to publication (wow, right!??!) that you can find in the abstract.
Got a couple files. This study was on an Orbitrap Velos running High-Low (FT for MS1 and CID ion trap MS/MS scans). It looks like 60k resolution at the MS1 (around 400m/z)
16 Kasım 2015 Pazartesi
Is offline fractionation hurting us more than its helping?
This is interesting -- and deserves more time than I can give it on a lunch break.
Its out of something called the Yates lab. Seems familiar, somehow...
The paper is called: "Off-Line Multidimentional LC and Auto Sampling Results in Sample Loss in LC/LS-MS/MS" and you can give it more time (appears open access! here)
Interesting, right?
Its out of something called the Yates lab. Seems familiar, somehow...
The paper is called: "Off-Line Multidimentional LC and Auto Sampling Results in Sample Loss in LC/LS-MS/MS" and you can give it more time (appears open access! here)
Interesting, right?
15 Kasım 2015 Pazar
Single protein matches with Mascot
This is an interesting post from this week on the Matrix Science blog. How to deal with single protein searches is Mascot.
14 Kasım 2015 Cumartesi
toxoMine -- use all omics to get rid of toxoplasmosis!
Toxoplasmosis is terrifying. Okay, a lot of diseases are....or I'm a coward. Not sure which. But stories of the this disease's subtle but powerful effects have been floating around the pop science scene for years. Malcolm Gladwell even mentions it in one of his great books.
Check out this article, titled: "Parasite makes men dumb, women sexy"
The problem with this disease is how highly evolved it is. Its right up there with Plasmodium in terms of complexity. And beating it...after millions of years of successfully existing in the mammalian population? That's gonna be real tough.
Maybe someone ought to take all the data we have on this thing from every Omics technology and make a great big awesome tool for digging through it!
TAAADAAAA!!! ToxoMine is exactly this tool. Interested in complicated diseases? This is a nice example of how we can align all the power these big -omics tools can give us!
Kaydol:
Yorumlar (Atom)
Popular Posts
-
A recent paper in PNAS makes the statement in the title "Protein Carbamylation is a Hallmark of Aging. You can find it here . They find...