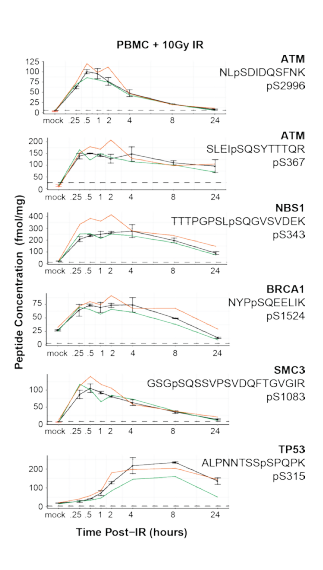
DNA damage response proceeds along a tightly controlled phosphorylation cascade. The main operators are very well studied and predictable enough that immuno assays are used in the clinic for these phosphorylation sites.
Could you quantify the entire cascade with a single LC-MS run? And could you do it with a high level of reproducibility? Sure looks like it!
In this paper from Jacob Kennedy et al., out of the Fred Hutch, they use a single step IMAC pull-down followed by MRMs and the data looks fantastic. (Max CV on phosphos of ~16%?!?!)
Do 150 Western blots? Or monitor all of this pathway in a single run? Makes the mass spec seem like a pretty cheap option, right? Now, being the resolution snob that I am, I would like to point out that the relatively small number of targets here, this is something that could easily be adapted to a Q Exactive. Again, the data looks seriously fantastic here, but if I was to improve this assay in any way I'd want to see my fragment ions in PRM +/- 1ppm.
1 Aralık 2015 Salı
Tardigrade genome is done!
How on earth do you guys do proteomics on unsequenced organisms? I know there's BICEPS, which relies on genomes from similar organisms and error tolerant searches, but...besides that?
Okay, here are some good examples. They finally did the tardigrade genome, and its reeeeaaaalllly weird. But if the genome is only just now done, how did:
These groups do the proteomics studies in 2010 and 2011?
Well, the 2010 group did 2D-gels and compare their IDs against the current known protein sequences and the group in 2011 focuses primarily on highly conserved heat shock proteins.
So, you do what you can with the proteins that are in the databases. Or you study something that doesn't change from organism to organism.
Why is this cool, other than the fact that tardigrades are frickin awesome? Cause this is a perfect example of a great meta-analysis project. Google Scholar pulls up 5 studies that appear to be at least partial proteomic analyses of these ridiculously cool organisms. Every single one of them was performed with imperfect genomic or protein databases. If I was looking to write a nice and reasonably high impact paper this weekend, I'd be downloading this genome here and seeing how many of these papers submitted data to Tranche and PRIDE that I can freely download and meta-analyze. Then maybe we'd know how these weird, indestructible things tick...or dance...or swim....
UPDATE 12/5/15: Ummm...okay, so maybe hold off on that meta-analysis... the work of this study above has been an explosion in the genomics community. It appears there might have been some errors. I LOVE SEEING SCIENCE IN ACTION!!! Maybe all this is real, and maybe not, but either way we'll be further ahead! Discussion on the controversy here.
29 Kasım 2015 Pazar
Proteomics in forensics!
Whoa!!
Despite what crime dramas might lead us to believe, forensics technologies still aren't perfect. A big hangup of the DNA evidence is that we have the same copy of DNA in every cell, so telling where the tissue samples came from is difficult/impossible.
Sounds like a job for proteomics!
In this cool paper from Sascha Dammeier et al., out of the Kohlbacher lab these researchers investigate using a proteomics approach to show that shotgun proteomics runs can tell you what tissue some evidence comes from. When your Materials and Methods section includes bullets and evidence bags, you've entered into interesting proteomics territory!
To make it even more interesting(!!) they did the proof-of-principle stuff on a cow organs subjected to blunt trama and then(!!!) they participated in a real crime investigation!!!
Okay, get this. A bullet passing through and organ carries enough protein for an Orbitrap XL to get a proteome signature good identify the organ!
Label free proteomics in Proteome Discoverer 2.0 is GO!
Got Proteome Discoverer 2.0?
Want to get a free upgrade that gives you awesome levels of label free proteomics capabilities?
I mentioned this before, but I just installed the nodes that are listed here on a fresh install of PD 2.0 and ran it and I'm blown away. These are really really super nice!
My recommendations (cause I don't know what I did wrong before):
1) Read the instructions
2) Download the nodes AND the processing and consensus workflows
3) Don't use a small file to test. Percolator needs to run for this to work. In PD 2.0 if you have less than 200 peptides going into Percolator it just turns off. Then the nodes can't work.
4) Revel in your new capabilities!!!!
(Look at this!!! I can find stuff in my runs that were not identified, but that were differentially (sp?) regulated in my two samples!!!! MAGIC!)
5) Remember that this is a free second party software. There is a nice mailing group you can sign up to for advice and news about the nodes. Your instrument vendor probably can't help you with these.
6) Do awesome label free proteomics!!!
27 Kasım 2015 Cuma
What genes can't humans live without?
This stuff has all been done on bacteria long ago, but these guys did this study in human cells. What genes (proteins!) are 100% essential for human cells to survive? Probably worth clarifying this statement -- what genes are 100% essential for human cells to survive in culture...cause thats the only way you're gonna pull this one off.
They come up with a list of 2,000. Wow, right? Depending on how you look at that, its either really big...or surprisingly small. I can't decide.
Are we surprised at all that a ton of them are proteins involved in protein glycosylation? Apparently that stuff is important.
Link here!
26 Kasım 2015 Perşembe
Turkey proteomics!
In celebration of my country's love of giant slow-cooked birds, I present you with this year's top hit on Google for "Turkey Proteomics!"
A EUPA meeting the third week of June in Istanbul?!?!? I LOVE the name of the meeting:
...Standardization and Interpretation of Proteomics.
25 Kasım 2015 Çarşamba
WGCNA -- Another way to post-process your quantitative data
I think its about time we start some concerted spy missions. The genomics people have all sorts of cool tools. Lets send proteomics people to genomics meetings and then just steal all their cool ideas, which is probably less like stealing because they give away virtually all of their algorithms.
Case in point? This WGCNA thing. Notice that its been around since 2008. So..around the time we were all getting our heads wrapped around target decoy searches, the genomics people were like "hey, lets do some unsupervised clustering of our thousands of quantitative changes and see what stands out in a hierarchical sense"
What's it do? It tries to find patterns in your complex data without you spending all day looking up genes that make sense. Its just pulling out common traits and clusters them with your sample(s) and type(s).
How's it do it? Well, its in R, so it does it in the ugliest way possible. Does it look like the screen of a Commodore 64? Yup! Then its likely R, he world's most powerful and utilized statistical software package!
Besides that? I have no idea. Google Scholar informs me that the initial paper describing this algorithm has been cited nearly 1,000 times. So, somebody has liked this paper. Maybe I'd even toy with it before trying to check the Stats.
You can visit the WGCNA website here. It has links to papers and full tutorials.
Shoutout to Alexis for mentioning this algorithm to me a second time and showing me cool clustering data from it so I'd remember to share with people!
Kaydol:
Yorumlar (Atom)
Popular Posts
-
A recent paper in PNAS makes the statement in the title "Protein Carbamylation is a Hallmark of Aging. You can find it here . They find...