21 Ekim 2015 Çarşamba
Skyline needs your help!
Ever wondered how the Skyline team has managed to create and support this awesome software? Turns out its got a grant in the background holding it up -- a grant that is up for competitive renewal. SO...this awesome free software that thousands of us are using (8,000 PCs fired up Skyline just last week!!!) might disappear (or become..gasp!..not free!) if this grant goes away.
If you're thinking "what can I possibly do to protect my access to this software before I finish this extremely large mug full of espresso shots?" you should click on this link.
It takes you to a place where you can download a draft letter of support that, if you get back to Brendan, will be attached to the R01 renewal application. The deadline for their application is coming up real fast, so time is of the essence.
20 Ekim 2015 Salı
TMT normalization based on variation is real important in clinical studies!
Any time we quantify anything we need to determine what is the priority for the measurement. Is it absolute quantification or is it lots of data points. This is always consistent, whether you're weighing out masses for buffers or doing global -omics quan.
We're absolutely no different. On Monday I worked with an awesome team developing an absolute quantification assay. One biomarker with heavy labeled spike-ins. One data point with precision and accuracy. But ONE measurment! Can we get that level of perfection with 16,000 measurements? Yeah...if you've got 10 years...(as technology improves...maybe 4 years?). And this will be measurement where we've reduced the level of technical variation.
Okay. So what happens when we look at something from a global level where the technical variation is high, but also the biological variation? I just learned that you can gain a lot of insight from this output by actually looking at the variation itself!
In this study in PLOS one, Evenlyne Maes et al., take a look at TMT labeled clinical samples and use statistical tools to normalize the samples using measurement variation. And all the sudden the data appears a lot more meaningful.
Variance normalization isn't a new concept in reporter ion based quantification. IsobariQ is a software a friend of mine has evaluated and really likes that was first showing off its algorithm for variance normalization in iTRAQ at HUPO in 2011. And there is an R package out there that does something similar and, for the life of me, I can never ever remember what it is called. Someone just asked me about it last week and I still haven't come up with the name.
What is cool about this paper is that we actually see the normalization working in clinical samples. Maybe someone else has done this, but I haven't seen it. If you show that something works well in your cool cancer cells that are grown at exactly 37C and get exactly 30% N2, or whatever, that is one thing (and awesome! and I'm not going to put it down ever. Bravo! I can't keep cells alive at all!)
But if you show me that your technique can extract more meaningful biological data out of people who walked into a clinic -- people who may have eaten 10 minutes ago, or 6 hours, or who have immunity to this virus or that or have any amount of the crazy differences each one of us walk around with every day? Thats gonna make me think we should take a look at what you did!
19 Ekim 2015 Pazartesi
Novor -- De novo peptide sequencing that's faster than your data acquisition
De novo sequencing has traditionally been a pain in the neck. If you are looking at low resolution MS/MS spectra there are often 10s, maybe 100s of possibilities out there to explain a given fragmentation spectra. High resolution accurate mass MS/MS spectra with fragments that are often only a few 100 parts per billion off in mass accuracy? That improves things a LOT! And that's why we're seeing things like de novo and wide mass accuracy windows making a resurgence.
You know what would be useful? A free (for academics...) super fast new de novo algorithm! And that is what Novor is. Novor makes some intelligent assumptions based on biological data (from spectral libraries) and can take a shortcut or two that other de novo algorithms can't. What you end up with is a crazy amount of sequencing speed without a loss in match certainty since its based on real data.
Novor has already been picked up in the new version of the awesome DeNovoGUI and is about to (or has, I forget...) appear in the super cool PeptideShaker.
If you're an academic you should check this out! If you aren't, you should check out their legal stuff or contact them to find out how you can use it.
You can read about Novor in this Open Access paper here.
And you can visit the RapidNovor website here.
You know what would be useful? A free (for academics...) super fast new de novo algorithm! And that is what Novor is. Novor makes some intelligent assumptions based on biological data (from spectral libraries) and can take a shortcut or two that other de novo algorithms can't. What you end up with is a crazy amount of sequencing speed without a loss in match certainty since its based on real data.
Novor has already been picked up in the new version of the awesome DeNovoGUI and is about to (or has, I forget...) appear in the super cool PeptideShaker.
If you're an academic you should check this out! If you aren't, you should check out their legal stuff or contact them to find out how you can use it.
You can read about Novor in this Open Access paper here.
And you can visit the RapidNovor website here.
18 Ekim 2015 Pazar
Happy birthday, SEQUEST!
Man, time flies when you're solving all the biological problems no one else can! SEQUEST is 20 years old now! To commemorate the fact that this awesome step forward for science will be old enough to have a drink with me next year JASMS has a special issue focusing on and around this accomplishment.
You can check the issue out here. Some articles are open access.
17 Ekim 2015 Cumartesi
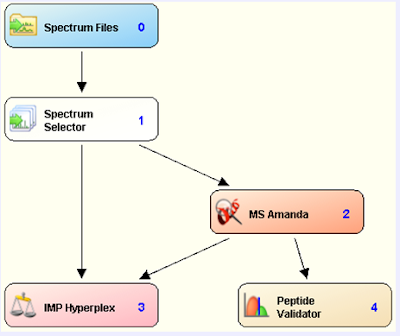
An improved TMT node for PD 1.4!
Pretty soon I'm going to be rambling on about all sorts of improvement in reporter ion quan due to improvements we'll be seeing in Proteome Discoverer 2.1.
In the meantime, however, here's a great new node for PD 1.4 that improved TMT quan for this software package.
How's it work? Well, it takes a bunch of things into account, including the reporter ion isotope distribution! I can't wait to give it a shot. Unfortunately, I think I've let my PD 1.4 licenses expire and I've got to get on that....
You can download this node along with installation and processing instructions from pd-nodes.org
16 Ekim 2015 Cuma
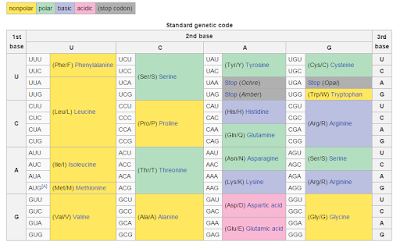
Amino acid composition and effects of mutations
That's pretty clear, isn't it? Just kidding, I have NO idea what that is, but its a figure from this paper, that does actually make a lot of sense.
Here is the gist of it: mutations are gonna happen. If they didn't evolution, to a large degree, wouldn't happen either (hugely complicated higher organism silly reproductive procedures aside). What Sahand Hormoz is proposing here is that the evolutionary pressure on essential proteins and their structures is to minimize the nasty effects of mutations is a mathematical certainty. That's a long sentence and I think the grammar could use some work.
Here, I've stolen the genetic coding table from this Wikipedia page. Say, you've got an essential leucine at this essential location. That's great! Cause many point mutations can happen to the section of the DNA and the protein won't change at all! You can change UUA to UUG or CUA and you're still gonna have leucine! Now, if it goes UAA...well you've got a truncated protein...so maybe this wasn't the best example.
There are also changes where we switch amino acids and it isn't all that bad either. For example, it would be better to have a mutation that changes a polar amino acid with a small functional group to another similar one, but changing to one with a huge non-polar, would be a whole lot worse.
From the genetic perspective, this makes all sorts of sense. What this guy did was show that you can calculate it with that Maths stuff to show that this wasn't accidental at all. This was evolutionary pressure over billions of years to find the best possible way to protect the proteins that life would end without. Really cool read, even if you don't know what any of the equations mean!
Hey, Maryland and D.C (and surrounding areas!) Wanna come to the NIH for this awesome workshop?
Hey! I know this is self-serving, but enrollment for the NIH Proteome Discoverer 2.0 workshop number 2 is still kinda low. I don't think its in danger of being cancelled, but if more attendees register then I'll be more confident we can have this thing.
The program is starting to take shape. The advanced concepts in the afternoon will definitely include:
How to combine quantitative whole protein and quantitative PTM (probably phosphoproteomics) into a single report AND
How to combine the results of multiple TMT and iTRAQ experiments.
We are still taking suggestions for other concepts to cover!
If you want to come, please register at this link: https://www.surveymonkey.com/r/?sm=PenWchsGrQuQZQmLH6odMzQoGuh4cDU%2fWF5ilEAy6oI%3d
Kaydol:
Yorumlar (Atom)
Popular Posts
-
A recent paper in PNAS makes the statement in the title "Protein Carbamylation is a Hallmark of Aging. You can find it here . They find...